Comprehensive Management of Cardiovascular Risk Factors for ...
AI Tool Pinpoints Genetic Mutations That Cause Disease
Google DeepMind has wielded its revolutionary protein-structure-prediction AI in the hunt for genetic mutations that cause disease.
A new tool based on the AlphaFold network can accurately predict which mutations in proteins are likely to cause health conditions — a challenge that limits the use of genomics in healthcare.
The AI network — called AlphaMissense — is a step forward, say researchers who are developing similar tools, but not necessarily a sea change. It is one of many techniques in development that aim to help researchers, and ultimately physicians, to 'interpret' people's genomes to find the cause of a disease. But tools such as AlphaMissense — which is described in a 19 September paper in Science — will need to undergo thorough testing before they are used in the clinic.
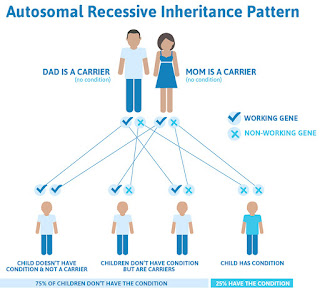
Many of the genetic mutations that directly cause a condition, such as those responsible for cystic fibrosis and sickle-cell disease, tend to change the amino acid sequence of the protein they encode. But researchers have observed only a few million of these single-letter 'missense mutations'. Of the more than 70 million possible in the human genome, only a sliver have been conclusively linked to disease, and most seem to have no ill effect on health.
So when researchers and doctors find a missense mutation they've never seen before, it can be difficult to know what to make of it. To help interpret such 'variants of unknown significance,' researchers have developed dozens of different computational tools that can predict whether a variant is likely to cause disease. AlphaMissense incorporates existing approaches to the problem, which are increasingly being addressed with machine learning.
Locating mutationsThe network is based on AlphaFold, which predicts a protein structure from an amino-acid sequence. But instead of determining the structural effects of a mutation — an open challenge in biology — AlphaMissense uses AlphaFold's 'intuition' about structure to identify where disease-causing mutations are likely to occur within a protein, Pushmeet Kohli, DeepMind's vice-president of Research and a study author, said at a press briefing.
AlphaMissense also incorporates a type of neural network inspired by large language models like ChatGPT that has been trained on millions of protein sequences instead of words, called a protein language model. These have proven adept at predicting protein structures and designing new proteins. They are useful for variant prediction because they have learned which sequences are plausible and which are not, Žiga Avsec, the DeepMind research scientist who co-led the study, told journalists.
DeepMind's network seems to outperform other computational tools at discerning variants known to cause disease from those that don't. It also does well at spotting problem variants identified in laboratory experiments that measure the effects of thousands of mutations at once. The researchers also used AlphaMissense to create a catalogue of every possible missense mutation in the human genome, determining that 57% are likely to be benign and that 32% may cause disease.
Clinical supportAlphaMissense is an advance over existing tools for predicting the effects of mutations, "but not a gigantic leap forward," says Arne Elofsson, a computational biologist at the University of Stockholm.
Its impact won't be as significant as AlphaFold, which ushered in a new era in computational biology, agrees Joseph Marsh, a computational biologist at the MRC Human Genetics Unit in Edinburgh, UK. "It's exciting. It's probably the best predictor we have right now. But will it be the best predictor in two or three years? There's a good chance it won't be."
Computational predictions currently have a minimal role in diagnosing genetic diseases, says Marsh, and recommendations from physicians' groups say that these tools should provide only supporting evidence in linking a mutation to a disease. AlphaMissense confidently classified a much larger proportion of missense mutations than have previous methods, says Avsec. "As these models get better than I think people will be more inclined to trust them."
Yana Bromberg, a bioinformatician at Emory University in Atlanta, Georgia, emphasizes that tools such as AlphaMissense must be rigorously evaluated — using good performance metrics — before ever being applied in the real-world.
For example, an exercise called the Critical Assessment of Genome Interpretation (CAGI) has benchmarked the performance of such prediction methods for years against experimental data that has not yet been released. "It's my worst nightmare to think of a doctor taking a prediction and running with it, as if it's a real thing, without evaluation by entities such as CAGI," Bromberg adds.
This article is reproduced with permission and was first published on September 19, 2023.
Researchers Uncover Why A Gene Mutant Causes Young Children To Have Strokes
α-smooth muscle actin localizes to the nucleus concurrently with smooth muscle cell differentiation. A, Immunoblot of fractionated protein lysates from SMCs explanted from WT mice shows αSMA localizes to the nucleus in SMCs, and both cytosolic and nuclear αSMA levels increase with TGF-β1 stimulation, while PDGF-BB stimulation does not affect nuclear accumulation of αSMA. B, Two-dimensional gel electrophoresis shows both αSMA and β-actin in the nucleus of SMCs. C, LtA treatment does not alter the ratio of nuclear to cytosolic αSMA on immunoblot. D–f, Immunostaining of isolated nuclei (d) shows increased nuclear αSMA after treatment with TGF-β1 or PDGF-BB, quantified in e, and confirms colocalization of αSMA (green) with the nuclear speckle marker sc-35 (red), quantified in f. For e, n = 59 untreated nuclei, 40 + TGF-β1 nuclei and 26 + PDGF-BB nuclei across four independent experiments. For f, n = 28 untreated nuclei, 19 + TGF-β1 nuclei, and 20 + PDGF-BB nuclei across three independent experiments. For e and f, significance was assessed by the Kruskal–Wallis test followed by Dunn's multiple-comparisons test. G, Quantitative RT–PCR shows exponential increases of SMC contractile gene expression during the time course of NEPC-to-SMC differentiation. H, Immunoblot of fractionated protein lysates taken at time points during the differentiation of NEPCs (day 0) to SMCs (day 12) shows early and dramatic accumulation of nuclear αSMA. β-actin is decreased in the nucleus of NEPCs. Time points match between g and h. Data shown are representative of at least three independent experiments. Quantifications of immunoblots can be found in Extended Data Fig. 1. Negative controls for immunostaining can be found in Supplementary Fig. 1. All data are presented as the mean ± s.D. Credit: Nature Cardiovascular Research (2023). DOI: 10.1038/s44161-023-00337-4A discovery of a mutation in the gene ACTA2 has given researchers, led by Dianna Milewicz, MD, Ph.D., with UTHealth Houston, insight into understanding the cause of a rare and progressive problem with arteries in the brain and a cause of strokes in young children, called moyamoya disease.
The findings were published today in Nature Cardiovascular Research.
Moyamoya disease is a condition where the arteries going through the neck and into the brain become blocked right when the arteries enter the brain. Moyamoya disease can lead to strokes and seizures. Children only a few months old can suffer a stroke due to the disease. Current treatments are limited to medications to reduce the risk of stroke and surgery to open or bypass blocked arteries.
"This disease is one of the major causes of stroke in children and nobody knows why this happens or why these arteries get clogged," said Milewicz, senior author of the study and professor and director of the Division of Medical Genetics at McGovern Medical School at UTHealth Houston. "These children don't smoke; they don't have hypertension and they don't have any of the other usual risk factors that cause strokes in adults."
Researchers identified that a change in the gene called ACTA2 caused children to have moyamoya disease and strokes starting shortly after birth, a condition called Smooth Muscle Dysfunction Syndrome, and have been working to figure out how and why this ACTA2 change causes moyamoya disease and strokes. Previous research led by Milewicz identified that ACTA2 mutations are the cause of Smooth Muscle Dysfunction Syndrome. In addition to moyamoya disease, this condition causes dysfunction of smooth muscle cells throughout the body.
ACTA2 is found in the smooth muscle cells, which line the arteries and allow them to contract to control blood pressure and flow. Using model systems, including cells from patients with the ACTA2 variant that causes moyamoya disease, Milewicz and her team found that a mutation in ACTA2 causes the cells in the walls of the arteries in the brain to not differentiate properly, an essential component of vascular development.
"We found a new job that the ACTA2 protein is supposed to do that the mutant version cannot: to help make differentiated smooth muscle cells that stay in the blood vessel and contract to regulate blood pressure," said Callie Kwartler, Ph.D., first author of the study and assistant professor in the Division of Medical Genetics at McGovern Medical School.
The result is that the cells with the ACTA2 variant continue to grow out of control and move into the inside of the artery, which may be the cause of blockages in the arteries.
"This is the first step into really understanding the cause of moyamoya disease," said Milewicz, the President George Bush Chair in Cardiovascular Medicine with McGovern Medical School. "This is a disorder that starts out in childhood, and children with Smooth Muscle Dysfunction Syndrome die from strokes. We are working to use the information to prevent strokes in these children."
Researchers will continue to focus on exploiting the mechanism of disease that they identified to find new treatment options for children with moyamoya disease.
More information: Callie S. Kwartler et al, Nuclear smooth muscle α-actin participates in vascular smooth muscle cell differentiation, Nature Cardiovascular Research (2023). DOI: 10.1038/s44161-023-00337-4
Citation: Researchers uncover why a gene mutant causes young children to have strokes (2023, September 28) retrieved 28 September 2023 from https://medicalxpress.Com/news/2023-09-uncover-gene-mutant-young-children.Html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no part may be reproduced without the written permission. The content is provided for information purposes only.
Advances In Gene Therapy For CLN2 Batten Disease
Effect of intracisternal administration of AAVrh.10hCLN2 vector on NHP safety, as scored from videotaped behavioral assessments at multiple time points. Four treated wild-type NHPs (African Green vervets; three males and one female) were assessed for behavior parameters at 4 time points: before study (Pre), day of administration (day 0), week 4, and week 8 (before necropsy). At indicated times, NHPs were videotaped. Results of 20 behavioral parameters (see Supplementary Table S1 for parameters assessed) for each NHP in the study are shown as sums. Linked data plots over time for each NHP (means of three reviewers, yellow, PBS [M]; green, PBS [M]; red, AAVrh.10hCLN2 [F]; blue, AAVrh.10hCLN2 [M]). Note: no NHP exhibited "Unhealthy" behaviors. NHP, nonhuman primate; PBS, phosphate buffered saline. Credit: Human Gene Therapy (2023). DOI: 10.1089/hum.2023.067A new study shows that delivery of gene therapy to correct the gene mutations that cause CLN2 disease, or Batten disease, directly into the cerebrospinal fluid (CSF) has potential therapeutic effects. The study, conducted in nonhuman primates, is published in the journal Human Gene Therapy.
CLN2 disease is a fatal, childhood autosomal recessive disorder cause by mutations in the CLN2 gene, which encodes tripeptidyl peptidase (TPP-1). In a prior study, the investigators, Ronald Crystal, MD, and Dolan Sondhi, Ph.D., from Weill Cornell Medical College, and co-authors, found that intraparenchymal administration of an adeno-associated virus (AAV) vector encoding human CLN2 slowed but did not stop disease progression.
The investigators concluded that this delivery route may have been insufficient to distribute the therapy throughout the central nervous system. Whereas in that study TPP-1 activity was >2X above controls in 30% of treated brains, in the current study, with delivery directly to the CSF, TPP-1 activity was >2X above controls in 50% and 41% of the brains of the two treated animals. CSF TPP-1 levels in treated animals were 43-62% of normal human levels.
"Batten disease is a profoundly tragic disorder in which children develop normally until around age 5, but then begin experiencing seizures, blindness and progressive loss of neurologic function," says Editor-in-Chief Terence R. Flotte, MD, Celia and Isaac Haidak Professor of Medical Education and Dean, Provost, and Executive Deputy Chancellor, University of Massachusetts Chan Medical School. "The advancement of this potential gene therapy could provide new hope to families with affected children."
More information: Bishnu P. De et al, Assessment of Safety and Biodistribution of AAVrh.10hCLN2 Following Intracisternal Administration in Nonhuman Primates for the Treatment of CLN2 Batten Disease, Human Gene Therapy (2023). DOI: 10.1089/hum.2023.067
Citation: Advances in gene therapy for CLN2 batten disease (2023, September 28) retrieved 28 September 2023 from https://medicalxpress.Com/news/2023-09-advances-gene-therapy-cln2-batten.Html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no part may be reproduced without the written permission. The content is provided for information purposes only.

Comments
Post a Comment